Structural work
Solving Tough Cases: Chaperone-Assisted Crystallography as a Robust Tool for Structure Determination of Nucleic Acids
Artur Laski1, Zoé Kürsteiner1, Kenny Jungfer2, Pascal Röthlisberger1, Andreas Gloger1, Jörg Scheuermann1, Martin Jinek2, Jonathan Hall1
Organization(s): 1: ETH Zürich, Switzerland; 2: University of Zürich, Switzerland
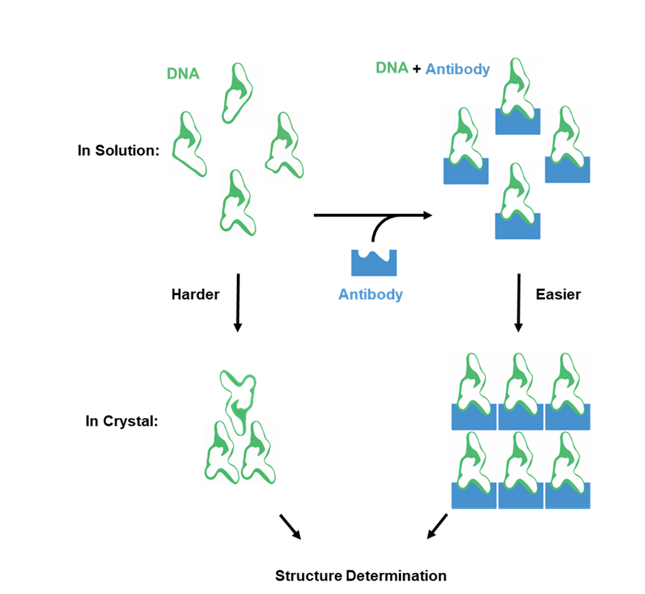
In the current era of rapid expansion in nucleic acid research, deciphering the structures of these molecules stands as the key step in determining their full application potential. X-ray crystallography emerges as an invaluable tool, providing atomic-level details of the spatial arrangement of atoms within crystallized nucleic acid molecules. This technique relies on capturing X-ray diffraction patterns resulting from the interaction of radiation waves with atoms in a three-dimensional, well-ordered molecular array known as a crystal. Computational reconstruction, based on the ordered arrangement of identical macromolecules in the crystal, yields accurate and high-resolution molecular structure data. However, despite recent advances, only a mere 6% of the ~179,000 unique X-ray structures in the PDB database consist of nucleic acid structures, with the majority contributed by protein structures. This disparity underscores persistent challenges in determining nucleic acid structures. Addressing these challenges requires developing new crystallization systems. One of the most successful approaches proves to be co-crystallization, utilizing protein chaperones such as synthetic antibody fragments (FABs) to aid in crystallization and phasing problems.
In this study, we adapted the RNA-binding FAB BL3-6, originally developed in the Joseph Piccirilli Lab1 , for the crystallization of DNAzymes. Through the re-engineering of plasmids and the establishment of robust production protocols for FAB BL3-6 in CHO cells, we have overcome production challenges associated with the original protocols. Subsequently, we synthesized large quantities of clean DNA-RNA mixmers engineered specifically to facilitate the crystallization of DNAzyme structures in their pre-catalytical state. Finally, we demonstrated that FAB BL3-6 binds to the engineered DNAzyme-RNA mixmers without compromising their catalytic functions. This comprehensive effort led to the determination of elusive structures of RNA-cleaving DNAzymes. We anticipate that these results will significantly enhance the crystallographic toolbox for understanding DNAzyme structures, with the potential to reveal new structures of other nucleic acids as well.
1. Koldobskaya, Y.; Duguid, E. M.; Shechner, D. M.; Suslov, N. B.; Ye, J.; Sidhu, S. S.; Bartel, D. P.; Koide, S.; Kossiakoff, A. A.; Piccirilli, J. A., A portable RNA sequence whose recognition by a synthetic antibody facilitates structural determination. Nat Struct Mol Biol 2011, 18 (1), 100-6.